Moore’s Law predicts that computing will dramatically increase in power and decrease in relative cost at an exponential pace. Although this principle mainly applies to computing hardware, DNA sequencing cost has followed a similar pattern for many years, approximately halving every two years. But since January 2008 there has been a break in that trend, with sequencing costs dropping much faster than the cost of processing data on computers. The cost of getting DNA data has never been cheaper, and it will continue to decrease.
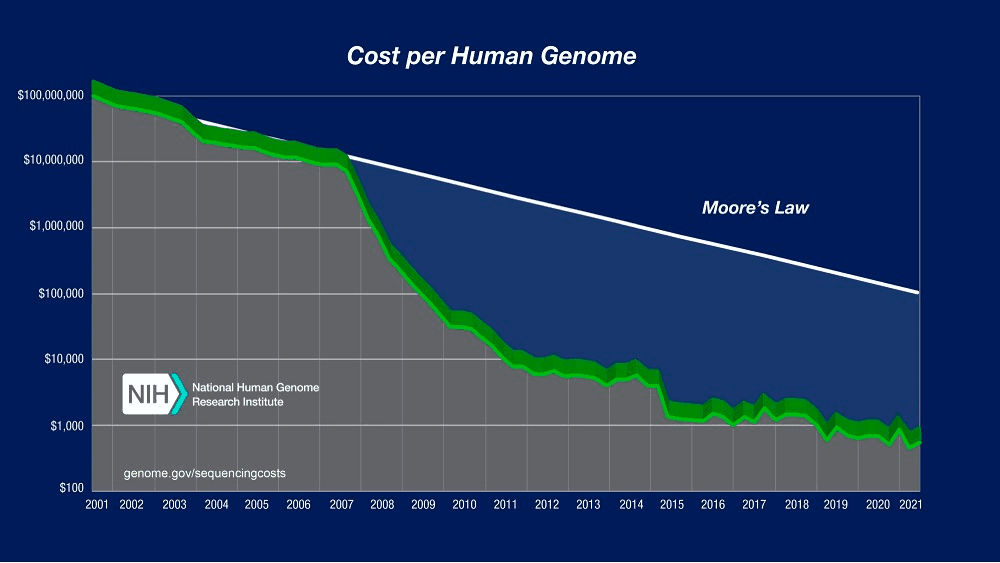
The result of this is that a data tsunami is coming into Life Sciences, with estimations that over 60 million patients will have their genome sequenced in a healthcare context by 2025, and that genomics research will generate between 2 and 40 exabytes of data within the next decade.
But it’s not just about data volume. Massive computing power has enabled researchers from the University of Illinois to develop a software to simulate a 2-billion-atom cell that metabolizes and grows like a living cell. Cell simulation provides insights into the physical and chemical processes that form the foundation of living cells, where fundamental behaviors emerge not because they were programmed in, but because the model contained the correct parameters and mechanisms.
Life Sciences is going through an accelerated transformation, and Machine Learning (ML) is responsible for it. On top of that, during the Covid-19 pandemic Life Sciences companies were forced to mobilize their resources to respond quickly to public health demands, which caused a spike of new computational methods and ways of thinking.
Applied Machine Learning in Life Sciences
Massive data volumes plus improved computation have eased the path to ML models that can solve new challenges in Life Sciences. From the big universe of potential applications, some that deserve special attention are:
- Improved diagnostics, since diagnosis is the most fundamental step in the treatment of any patient.
- Drug discovery, as the biggest goal of the Life Sciences industry is to advance the research and innovation for new products and treatments.
Improving diagnostics with Computer Vision
Computer Vision (CV) focuses on image and video understanding, involving tasks such as object detection, image classification, and segmentation. In Life Sciences, CV models fed with medical imaging (e.g. MRI, X-rays, etc) can assist in the visualization of cells, tissues and organs to enable a more accurate diagnosis, helping to identify any issues or abnormalities.
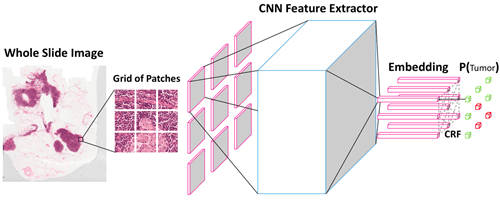
There’s huge versatility within imaging sources, since computed tomography (CT) scans and magnetic resonance imaging (MRI) are capable of generating 3D image data, while digital microscopy can generate terabytes of whole slide image (WSI) of tissue specimens.
One example is the UC San Diego Health, which applies CV models to quickly detect pneumonia through X-rays imagery, which are cheaper and faster than other methods. But adding Transfer Learning (TL) to these types of problems can increase the performance and accuracy of a diagnostic approach that medical professionals can easily use as an auxiliary tool. In CV, embeddings are often used as a way to translate knowledge between different contexts, allowing us to exploit pre-trained models like VGG, AlexNet or ResNet.
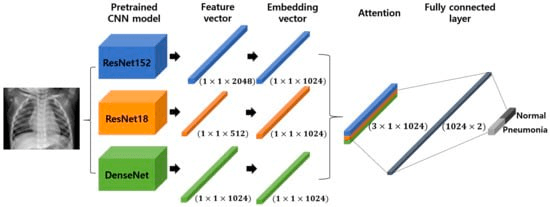
Due to the need for large datasets to train and tune Deep Learning architectures for CV which are not available for medical images, TL coupled with embeddings can be used to achieve tasks that go from ocular disease recognition to cancer detection.
To solve CV challenges, the classic approach has been to use TL with pre-trained convolutional neural networks (CNNs) on natural images (e.g., ResNet), tuned on medical images. Today, due to their powerful TL abilities, pre-trained Transformers (which are self-attention-based models) are becoming standard models to improve results on CV tasks.
Drug discovery
Drug discovery is the process of finding new or existing molecules with specific chemical properties for the treatment of diseases. Since this has traditionally been an extremely long and expensive process, modern predictive models based on ML have gained popularity for their potential to drastically reduce costs and research times.
ML can be used the in the drug discovery processes to:
- Discover structure patterns: to study the surface properties, molecular volumes or molecular interactions.
- Identify behavior influences: to relate the orientation of the molecule to its characteristics.
- Anticipate characteristics: to develop models capable of predicting the behavior of a molecule in accordance with its design.
- Improve drug designs: to get better results and design better medicines while reducing costs.
This is what companies like Sanofi are doing in order to reduce the sheer number of compounds they need to synthesize in the real world by doing much of the analysis on a computer.
How do you represent molecules in the data space? From the several representation methods available, embedding methods like Mol2Vec have emerged as novel approaches to learn high-dimensional representations of molecular substructures. Inspired by word embedding techniques known as Word2Vec, Mol2Vec encodes molecules into a set of vectors that represent similar substructures in proximity to one another in the vector space. In a Natural Language Processing (NLP) analogous fashion, molecules are considered as sentences and substructures as words.
But molecules can also be represented as graphs. Graphs are a ubiquitous data structure, employed extensively within computer science and related fields. Social networks, molecular graph structures, biological protein-protein networks, recommender systems — all of these domains and many more can be readily modeled as graphs, which capture interactions (i.e., edges) between individual units (i.e., nodes).
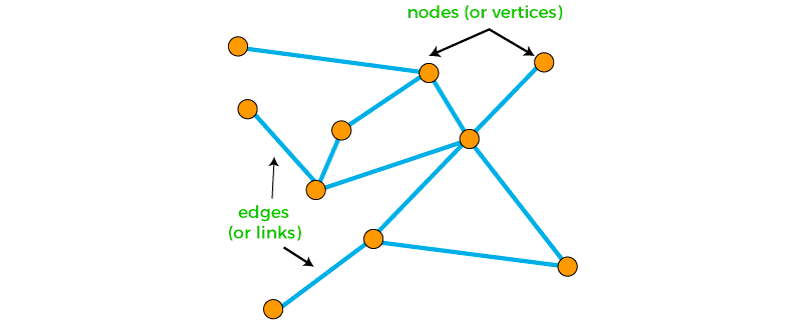
Intuitively, one could imagine treating the atoms in a molecule as nodes and the bonds as edges. Nodes, edges, subgraphs or entire graphs can be embedded into low-dimensional vectors that summarize the graph position and the structure of their local graph neighborhood. These low-dimensional embeddings can be viewed as encoding or projecting graph information into a latent space, where geometric relations in this latent space correspond to interactions in the original graph.
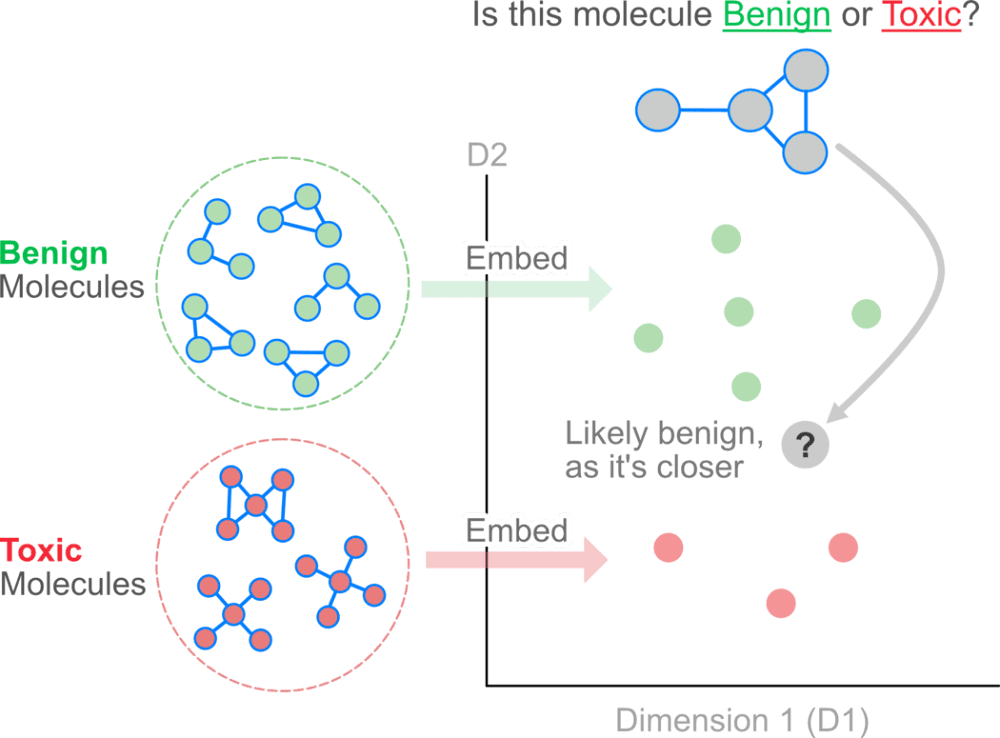
The idea behind using graph embeddings is to create insights that are not directly evident by looking at the explicit relationships between nodes.
The future
The data explosion and initiatives in Life Sciences have the potential to reshape the future of the industry and of patient care, as we witness how ML methods can do amazing things if you give them enough data. Just look at what DeepMind announced only some weeks ago, releasing the predicted structures for almost every protein known to science (over 200 million structures in total), using its AI AlphaFold 2.
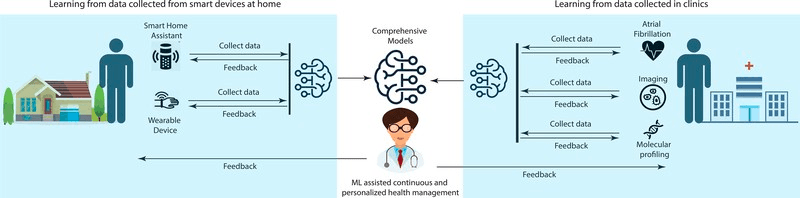
But it’s both the volume and diversity of data that force us to rethink how to solve problems in Life Sciences with ML. Life Sciences is demanding us to integrate all sorts of different data types to reach better results, while giving us a glimpse of what’s coming next for all industries: a multimodal future.
Consider Electronic Health Records (EHRs), which offer an efficient way to maintain patient information and are becoming more and more widely used by healthcare providers around the world. EHRs can include data that go from images, clinical notes, medication lists, vital signs, to demographic information, which can provide deep insights of a patient’s condition if integrated in an effective manner.
Efforts to integrate these data types are already ongoing, and multimodal ML models trained on numerous types of data could help health professionals to screen patients at risk of developing diseases like cancer more accurately. This is what Harvard University is researching, by training ML models with microscopic views of cell tissues from whole-slide images (WSIs) and text-based genomics data. Just imagine what other challenges can be faced when integrating image, sequential, text, 3D, graph and other types of data into the same information space.
ML will transform Life Sciences, and allow us to dream big in solving some of the most transcendental challenges for humanity, like eliminating aging or hyper-personalized healthcare. This is such a powerful Artificial Intelligence (AI) application, that the UK government has established it as a Strategic Grand Challenge, and people like Vitalik Buterin (the creator of the cryptocurrency Ethereum) and Jeff Bezos (founder of Amazon) invested part of their fortune in this idea. From personalized medicine to democratizing healthcare in developing regions, applied ML in Life Sciences can deeply change our lives.
Was this article helpful?